Sometimes the pathogens that infect plants also affect people - through our pocketbooks. This is why plant pathologist Erica Goss, a University of Florida professor with the Institute of Food and Agricultural Sciences, spends a lot of time studying microbes that infect tomatoes, peppers, and strawberries. These are the state’s top three grossing vegetable and berry crops, according to the Florida Department of Agriculture and Consumer Services, and a lot of microbes like them just as much as we do.
In 2019, the Florida tomato industry produced 39% of the US market for fresh tomatoes, worth about $426 million, while Florida’s fresh pepper crop was valued at $235 million. This is why growers are worried about pathogens that cause dark dried-out-looking blemishes on tomatoes and peppers.
 Tomatoes with brown spots caused by X. perforans fetch lower prices in fresh market sales. (Credit: Gary Vallad)
Tomatoes with brown spots caused by X. perforans fetch lower prices in fresh market sales. (Credit: Gary Vallad)
“We lump several different bacterial species under this disease called bacterial spot of tomato and pepper,” said Goss, who is also a faculty member of the UF Emerging Pathogens Institute. “But different species and even individual strains have different plant hosts and geographic ranges.”
The disease has been known to scientists for 100 years and there are four Xanthomonas lineages that cause a bacterial spot of tomato and pepper. X. perforans is the one found on tomatoes all over the Sunshine State today.
Goss frequently collaborates on her research into bacterial spots of tomato and pepper with two UF professors: Jeffrey B. Jones, a bacteriologist, is a distinguished professor in IFAS; and Gary Vallad, is a plant pathologist and bacteriologist at the UF/IFAS Gulf Coast Research and Education Center. Jones has studied Xanthomonas for more than three decades and Vallad works closely with Florida tomato growers and has expertise in plant disease epidemiology and disease management. The trio has been awarded grants together (from USDA and the Florida Tomato Committee) and in their most recent work, they applied some unusual methods to better understand the genetic diversity of this microbe and how it spreads to new host plants.
Tracing X. perforans through fields and production
When a tomato or pepper plant becomes infected with X. perforans, it develops spots on its leaves or fruits. The infected leaves may become spotted, or wither and die. When this happens, the fruits can develop sunscald, but the fruits can also become infected if the pathogen enters through the blossom, leading to malformed or spotted tomatoes or peppers. Either way, they fetch less money but are harmless to eat.
But how do the plants become infected? It usually starts with infected seeds, and with an international supply network, this spells trouble.
Few people are aware that the production of tomatoes is international,” Goss said. “You may grow a tomato in Florida, but it could have started as a seed in Asia. These are all hybrid seeds.”
Goss, Jones, and Vallad’s teamwork has shown there are many pathways in addition to seeds. The microbes could sit in greenhouses and then spread to the seedlings as they grow. Or, most greenhouses have an overhead irrigation system and X. perforans could hitch a ride on water droplets that splash from an infected leaf to an uninfected one. Or it may sit in soil in the growing fields or have an undetected reservoir in other plants. Somehow it colonizes fields once the tomato and pepper plants are growing.
Plant-to-plant spread can happen in the fields too. Air with high humidity or splashes from one plant to the next due to wind-driven rain or sprinkler irrigation can foster spread. Leaves of two different plants might also touch and foster plant-to-plant spread.
While growers can sometimes use copper to control the disease, X. perforans has quickly picked up close to 100% resistance via genes on plasmids. Plasmids are exchangeable packets that bacteria use to swap beneficial DNA. “Once it is copper resistant, there are not many options for reducing this disease in the field,” Goss said.
Where X. perforans is found
X. perforans is prolific. It occurs along the East Coast, but also in central Mexico, eastern Canada, southeastern Brazil, Sicily, Iran, Ethiopia, Tanzania, Thailand, Indonesia, Australia, Taiwan, and South Korea. Its broad global distribution is a reflection of the global trade networks for seeds.
“Why does it seem like this pathogen is getting to be a big problem in recent years? Is that pathogen evolution, or are new strains coming in from somewhere else? Or has something else about production changed?” Goss asked rhetorically, explaining the questions that guide her research. “How do we reduce the disease in the field? How is it getting out there and spreading?"
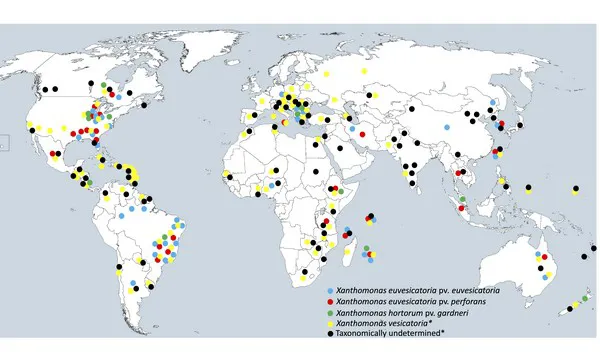 Map showing global distribution of Xanthomonas species and pathogenic variants. (Map courtesy of Erica Goss)
Map showing global distribution of Xanthomonas species and pathogenic variants. (Map courtesy of Erica Goss)
Understanding how and why X. perforans changes genetically
Goss uses molecular techniques, such as whole genome DNA sequencing, to detect genetic variation in X. perforans and changes over time.
“The X. perforans we’ve been studying in Florida seems remarkably genetically diverse,” Goss says. “There are these lineages that we find that are very different from one another, but then we also have a lot of variation within those. It’s clearly evolving very quickly.”
One of her current projects is supported by a US Department of Agriculture grant to detect genetic changes and variation in tomato fields. She plans to look at mutation rates and monitor for the movement of plasmids between host plants. This will give insight into how the bacteria is diversifying within Florida after being introduced at some point from the production system.
Other recent molecular work Goss, Jones, and Vallad have published this year characterizes the state of X. perforans in Florida. It details how X. euvesicatoria used to be the cause of bacterial spot of tomato and pepper in Florida, but was outcompeted by X. perforans in the early 1990s. Her team collected 585 Xanthomonas strains from 70 tomato fields in Florida and found that the strains were genetically diverse and that nearly half produced a compound capable of killing X. euvesicatoria.
In a different paper, published this spring, the team reported the use of unusual methods to test how a specific gene gave X. perforans an advantage in the field. They wanted to test how an effector gene, xopJ2, influenced the microbes’ fitness. Others had hypothesized the gene conferred an advantage, but no one had direct evidence of what it did.

Tomato plant leaves showing brown, dried-out spots characteristic of infection with a common pathogen, Xanthomonas perforans. (Credit: Gary Vallad)
Goss’ collaborators used lab techniques to engineer a mutant form of xopJ2 that was silenced. They then tracked two main groups of X. perforans in the field: one that had the natural, functional xopJ2 gene, and one that had the silenced version.
“This was like a mark-release-recapture study, but to look at the relative spread of the different bacteria,” Goss said, referring to a type of study design used by ecologists. It helps to estimate a population’s size in cases where it can be impractical to count every organism, in this case, to study the effect of the gene on the population of the pathogen.
They found that the microbes with the functional gene moved three times as fast in the field compared with the microbes that had the silenced, mutated version. Such a powerful advantage in the ability to move, combined with the deadly compound that can kill competing types of Xanthomonas, may explain how this lineage was able to rapidly replace the previous X. euvesicatoria lineage in Florida.
“But we noticed several years ago that one of the lineages of strains we were studying had lost the ability to make this compound,” Goss said. “It makes it a better competitor on the leaf when X. euvesicatoria is present, but in its absence, we hypothesize that it’s a waste of energy to make, it’s too costly. So they stopped making it.”
In a third paper, the team explored how the tomato production system influences genetic diversity in X. perforans. They found that, when growers produced their own seeds, the strains of X. perforans on their plants tended to be less diverse and genetically distinct. In contrast, growers that used multiple sources for seeds or seedlings were mixing pots for these microbes: they had the most genetically distinct and diverse strains of X. perforans.
Looking forward
Goss said findings from this work could potentially be used to engineer genes resistant to X. perforans that can be bred into tomato and pepper plants. The work is costly and slow, but she is collaborating with researchers at the Gulf Coast Research Center to identify multiple resistance genes.
“It may not work against 100% of the pathogen’s population, but even if it works against 30-40%, it's better than no resistance,” Goss said.
She is also investigating pathogens that affect strawberries and cacao, and cases where pathogens have suppressed invasive grasses.
“My goal is to better understand how a plant pathogen emerges,” Goss said. “What are the ecological and evolutionary mechanisms behind a new or worse disease problem? How is the pathogen evolving or are new versions coming in?”
Understanding these processes will lead to a more refined understanding of what helps promote a destructive plant pathogen and its success. Knowledge like this will help crop scientists and growers in years to come, to ensure both crop values and a healthy food supply.
For more information:
Emerging Pathogens Institute of the University of Florida
www.epi.ufl.edu
